HOME | DD
 Albertonykus — Vertebrata Phylogeny
Albertonykus — Vertebrata Phylogeny
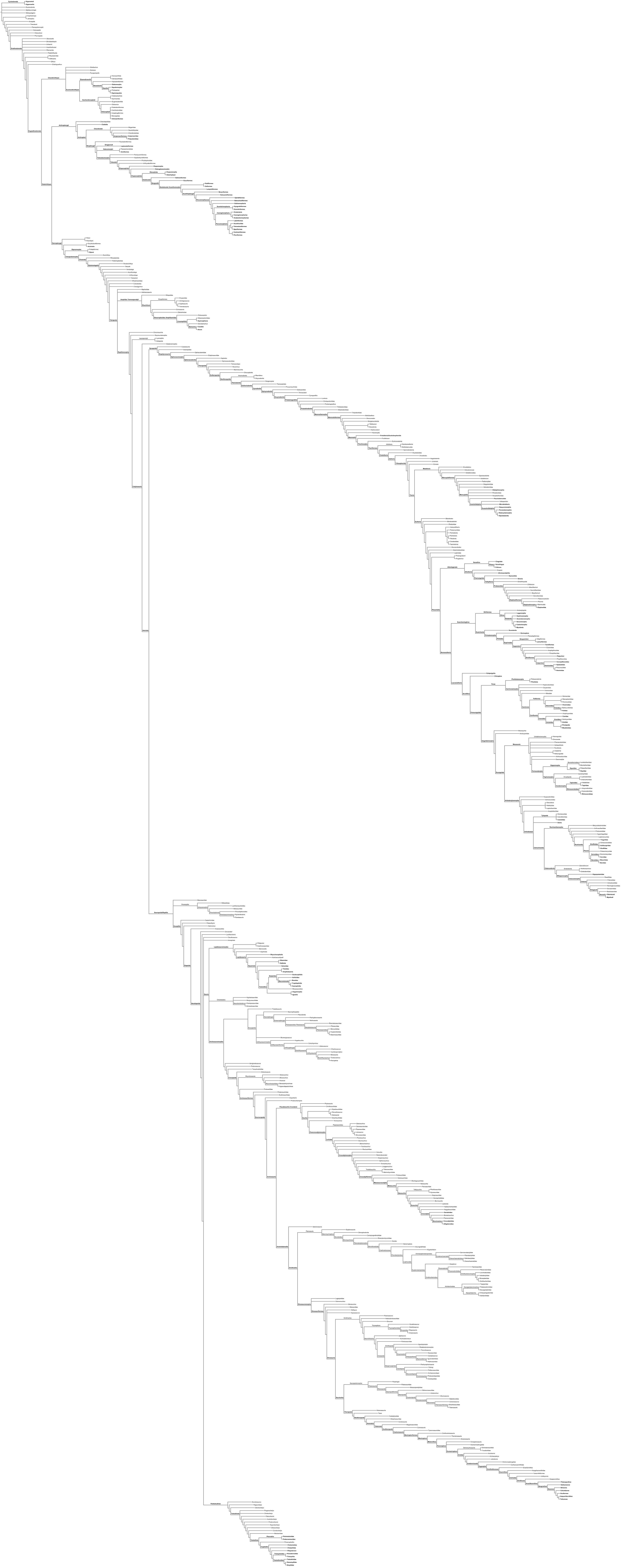
#birds #amphibians #dinosaurs #fish #mammals #reptiles #vertebrates #phylogeny #synapsids
Published: 2017-05-15 01:01:35 +0000 UTC; Views: 2517; Favourites: 53; Downloads: 78
Redirect to original
Description
I said I wouldn't be updating the Big Vertebrate Tree , but so long as Drs. Holtz and Merck continue making their course materials available publicly, there's nothing stopping me from updating just its general topology. Taxa in bold have extant representatives. I have graduated from UMD, and do not have inside info on why specific topologies may have been favored over alternative hypotheses.Edit: No (intentional) changes to the topology here, but I discovered the program TreeGraph for manipulating phylogenetic trees and decided to redo this tree using it. The aesthetics and readability have improved a good deal, I think.
Related content
Comments: 62

How do researchers work out these phylogenetic relationships?
👍: 0 ⏩: 1

Here are some lecture notes from another one of Holtz's classes that might be helpful. (In particular, the section titled "Phylogenetic Analysis: Methods".) In practice, phylogeneticists often consider hundreds of characteristics (or even thousands, if using molecular data) at once, so the process is usually done using computer software rather than by hand, but this should help you understand the principles involved.
Additionally, there are a few other methods of phylogenetic analysis (such as maximum likelihood and Bayesian) besides the one presented here, which is known as parsimony. However, parsimony is most frequently taught at an undergraduate level because it is the most intuitive and is still widely used in paleontological research. The main difference between the different methods is in the underlying algorithms used to determine what phylogenetic topology best fits the available data.
👍: 0 ⏩: 1

OK. So, stuff like TNT?
👍: 0 ⏩: 1

TNT is one of the programs commonly used to conduct phylogenetic analysis by parsimony, yes. Another one is PAUP.
👍: 0 ⏩: 1

So which of the methods is considered most reliable? (Bayesian vs parsimony vs maximum likelihood), or would you use multiple ones simultaneously to corroborate phylogenies?
👍: 0 ⏩: 1

That is an area of active debate. Some recent studies have concluded that Bayesian is generally more accurate at reconstructing the topology of phylogenies than parsimony, at the cost of lower resolution (that is to say, the results of Bayesian analyses are likely to have more parts of the tree where the branching order is uncertain). To my knowledge, among phylogeneticists there aren't yet many firm opinions regarding the accuracy of Bayesian versus maximum likelihood, but Bayesian has the advantage of being able to take into account more complex models of evolution.
However, some researchers do choose to use several methods for corroboration (e.g.: Brusatte and Carr, 2016 ), and there are particular situations that may favor specific methods over others. For instance, parsimony is still widely used for morphological data partly because Bayesian analysis has trouble working with continuous characters. (Continuous characters are characters not coded as discrete states such as 0 vs. 1 vs. 2, but instead as a range of states, e.g.: 1-3.) Additionally, Bayesian analyses take a very long time to process, so even phylogeneticists who prefer Bayesian might use parsimony if they just needed a "quick and dirty" result (e.g.: to check that their data matrix has a reasonable phylogenetic signal).
👍: 0 ⏩: 1

Thanks. So, it's still somewhat in the works, I take it?
👍: 0 ⏩: 1

Ah, ok. I'm currently reading Richard Dawkins's* "The Ancestor's Tale", which granted is more for the laypublic, is a pretty good and informative read, and it does discuss phylogenies and how cladograms/phylograms are set up in some depth. In future, though, I hope to learn a bit more than that.
*whom I affectionately call "Dicky D."
👍: 0 ⏩: 1

I quite enjoyed The Ancestor's Tale. Good overview of many of the major topics in evolutionary biology presented in a unique way.
👍: 0 ⏩: 1
<= Prev |
























